- Home
- Products
- Pathway
- Support
- Contact Us

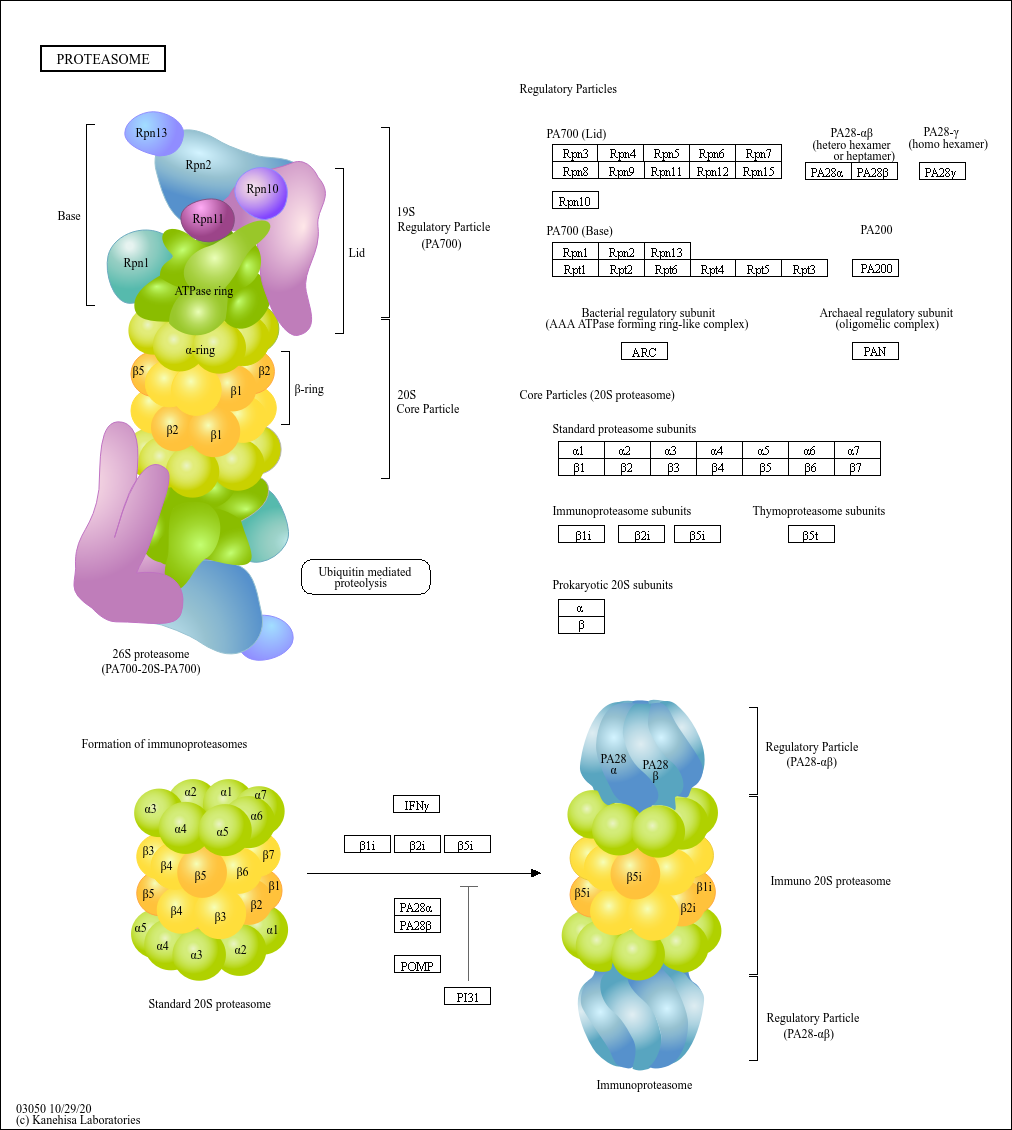
Proteasome
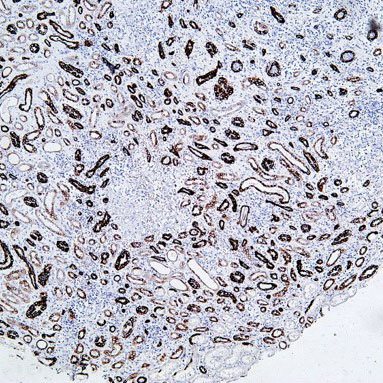
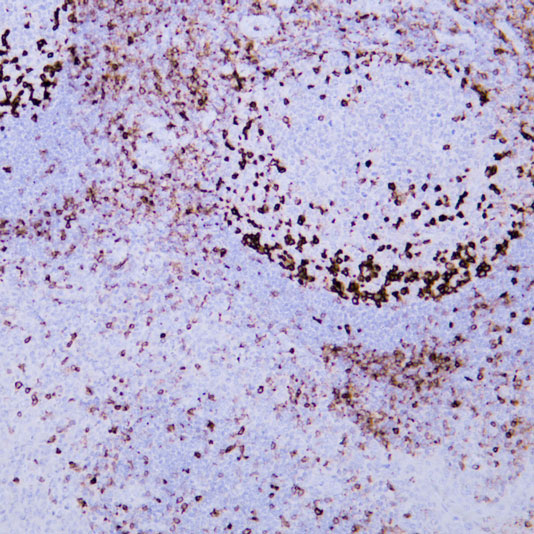
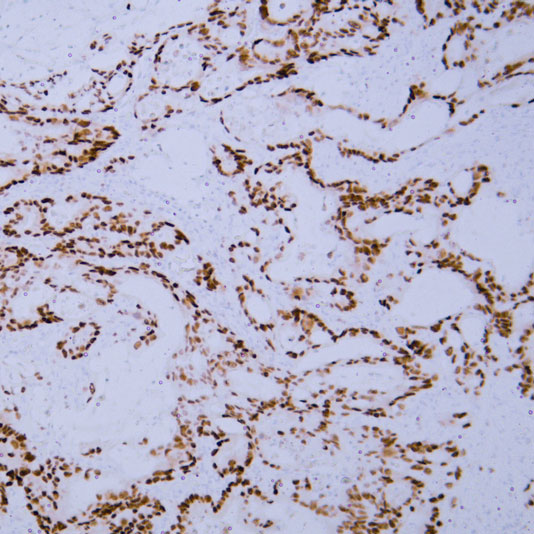
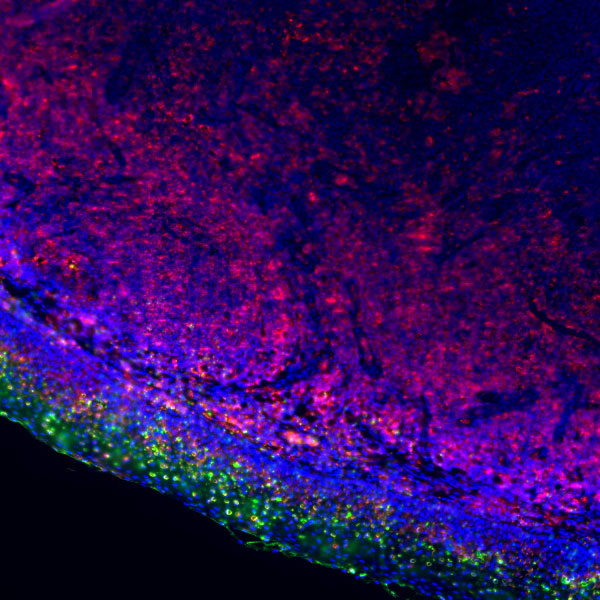
Core of basic research: Focuses on the molecular mechanism by which the 26S proteasome specifically degrades ubiquitinated proteins, a key process for regulating cellular protein homeostasis. The 26S proteasome consists of a 19S regulatory particle and a 20S catalytic particle: The 19S particle recognizes and binds to target proteins labeled with polyubiquitin chains, unfolds the protein conformation, and transports it to the 20S catalytic particle. The 20S particle contains trypsin-like, chymotrypsin-like, and caspase-like active sites, degrading proteins into short peptides (subsequently hydrolyzed into amino acids for recycling). Degradation substrates include misfolded proteins, cell cycle regulatory proteins, transcription factors, and oncoproteins, participating in cell cycle progression, signal transduction, and apoptosis regulation. Research focuses include the substrate specificity of proteasomal degradation, the synergistic effect between the ubiquitin-proteasome system and other degradation pathways (e.g., autophagy), and the association of pathway abnormalities with tumors (proteasome inhibitors used for tumor treatment) and inflammation.
Core key proteins: 26S proteasome (19S regulatory particle: subunits Rpn1, Rpn10, Rpt1-Rpt6; 20S catalytic particle: subunits β1, β2, β5), ubiquitin-binding proteins (recognize ubiquitinated substrates), ATPase subunits (Rpt1-Rpt6, provide energy for unfolding), proteasome activators (PA28, PA200), polyubiquitin chains (label degradation substrates), cyclins (e.g., Cyclin), oncoproteins (e.g., c-Myc), misfolded proteins.
Core key proteins: 26S proteasome (19S regulatory particle: subunits Rpn1, Rpn10, Rpt1-Rpt6; 20S catalytic particle: subunits β1, β2, β5), ubiquitin-binding proteins (recognize ubiquitinated substrates), ATPase subunits (Rpt1-Rpt6, provide energy for unfolding), proteasome activators (PA28, PA200), polyubiquitin chains (label degradation substrates), cyclins (e.g., Cyclin), oncoproteins (e.g., c-Myc), misfolded proteins.
Product list
-
{{item.title}}{{item.react}}{{item.applicat}}
Product list
Product name
Reactivity
Application
Related Resource Links
Related Promotional Journal Downloads

Explore Our Recommended Popular Products
More products
30,000+ high- quality products available online
Primary Antibodies, Secondary Antibodies, mIHC Kits, ELISA Kits, Proteins, Molecular Biology Products,Cell Lines,Reagents ...
Contact Us