- Home
- Products
- Pathway
- Support
- Contact Us

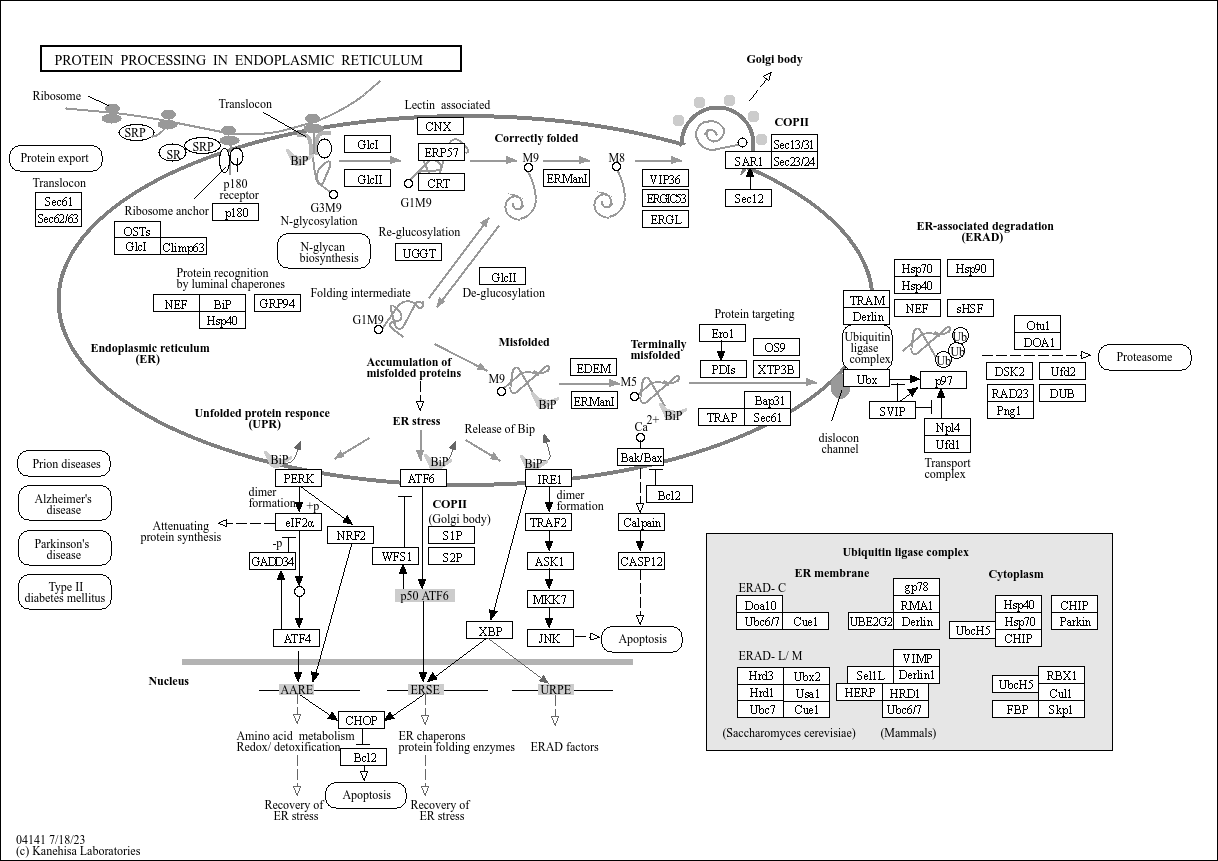
Protein processing in endoplasmic reticulum
Core of basic research: Clarifies the molecular mechanisms of protein folding, modification, and sorting in the endoplasmic reticulum (ER), a core process for the maturation and quality control of secretory proteins and membrane proteins. After nascent peptide chains enter the ER, they fold correctly with the assistance of chaperones (e.g., BiP, Calnexin, Calreticulin). Meanwhile, post-translational modifications occur: N-glycosylation (catalyzed by oligosaccharyltransferase), disulfide bond formation (catalyzed by PDI), and signal peptide cleavage (catalyzed by signal peptidase). Misfolded proteins are recognized via the ER-associated degradation (ERAD) pathway, transported to the cytoplasm, and degraded by the ubiquitin-proteasome system. If abnormal proteins accumulate excessively, the unfolded protein response (UPR) is triggered to regulate ER function and maintain cellular homeostasis. Research focuses include the molecular mechanism of protein folding, the functional specificity of glycosylation modification, the regulatory logic of ERAD and UPR pathways, and the association of pathway abnormalities with neurodegenerative diseases (e.g., Alzheimer's disease) and metabolic diseases (e.g., diabetes).
Core key proteins: Chaperones (BiP/GRP78, Calnexin, Calreticulin), oligosaccharyltransferase (OST, catalyzes N-glycosylation), protein disulfide isomerase (PDI, catalyzes disulfide bond formation), signal peptidase (cleaves signal peptides), ERAD-related proteins (Derlin-1, HRD1, p97/VCP), UPR pathway proteins (sensors IRE1, PERK, ATF6), ubiquitinases (associated with ubiquitination of abnormal proteins), secretory protein/membrane protein precursors.
Core key proteins: Chaperones (BiP/GRP78, Calnexin, Calreticulin), oligosaccharyltransferase (OST, catalyzes N-glycosylation), protein disulfide isomerase (PDI, catalyzes disulfide bond formation), signal peptidase (cleaves signal peptides), ERAD-related proteins (Derlin-1, HRD1, p97/VCP), UPR pathway proteins (sensors IRE1, PERK, ATF6), ubiquitinases (associated with ubiquitination of abnormal proteins), secretory protein/membrane protein precursors.
Product list
-
{{item.title}}{{item.react}}{{item.applicat}}
Product list
Product name
Reactivity
Application
Related Resource Links
Related Promotional Journal Downloads

Explore Our Recommended Popular Products
More products
30,000+ high- quality products available online
Primary Antibodies, Secondary Antibodies, mIHC Kits, ELISA Kits, Proteins, Molecular Biology Products,Cell Lines,Reagents ...
Contact Us