- Home
- Products
- Pathway
- Support
- Contact Us

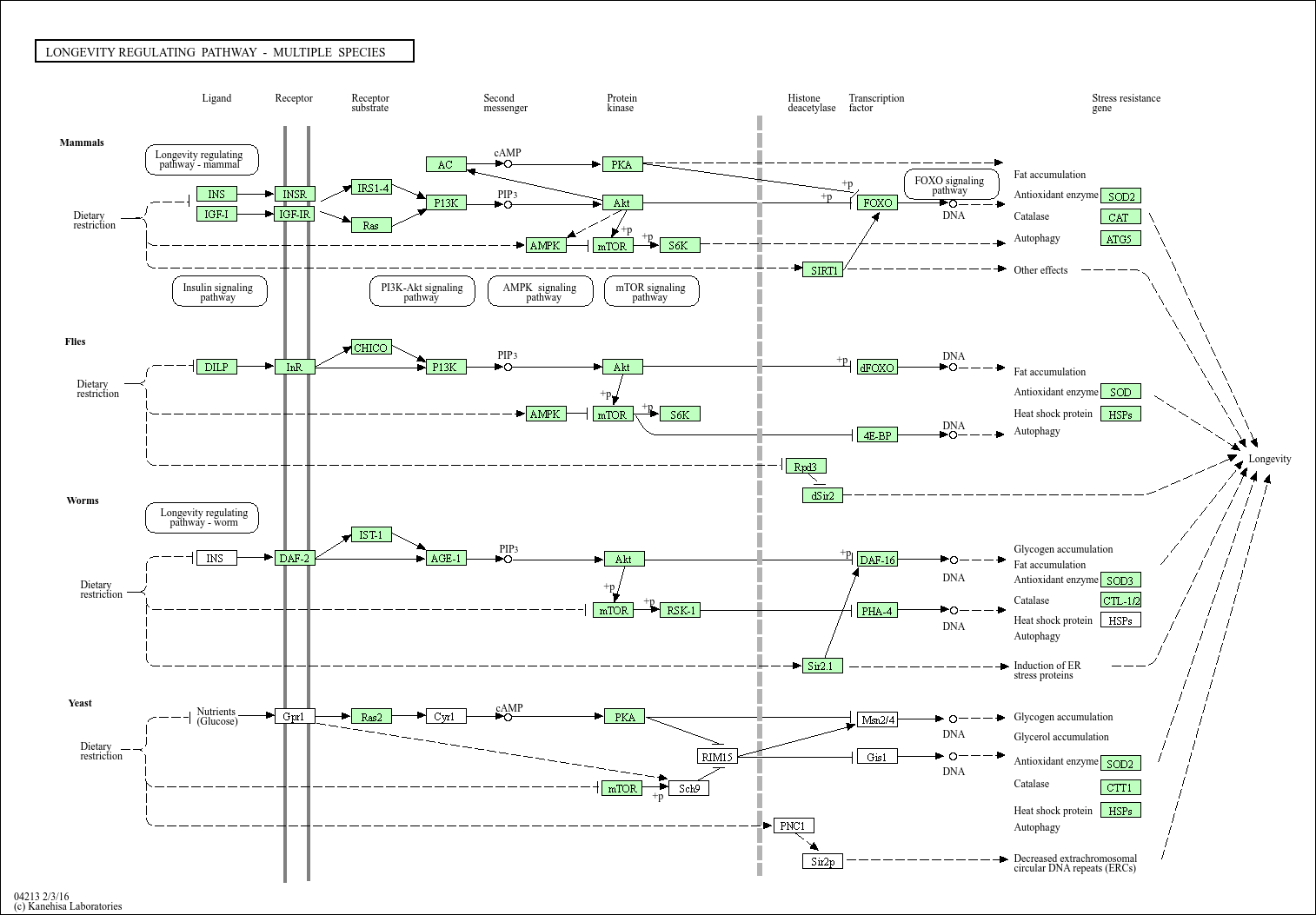
Longevity regulating pathway - multiple species
Core of basic research: Explores conserved longevity mechanisms across yeast, nematodes, Drosophila, and mammals, revealing evolutionarily shared aging logic. In nematodes, DAF-2 (insulin receptor homolog) mutations activate DAF-16 (FOXO homolog), upregulating stress resistance genes to extend lifespan. In yeast, SIR2 (SIRT1 homolog) extends lifespan via chromatin silencing and DNA repair. In Drosophila, inhibiting TOR or insulin/IGF-1 signaling significantly prolongs lifespan. In mammals, these pathways are conserved, and FOXO3 polymorphisms are closely linked to human longevity. Research focuses on conservation/species-specificity of longevity pathways, functional conservation of key genes (FOXO, SIRT1, TOR), environmental regulation (nutrition, temperature, exercise), and translational application of cross-species mechanisms.
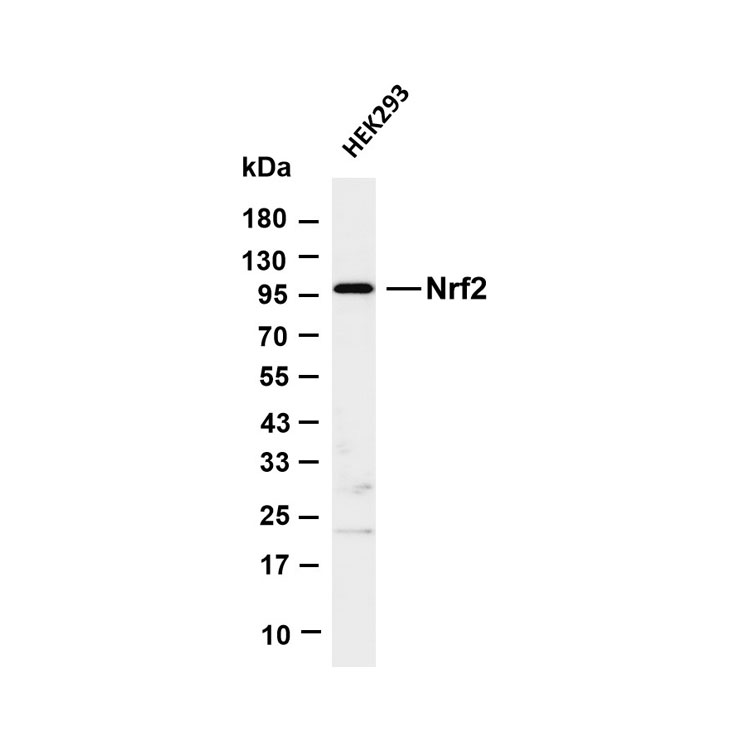
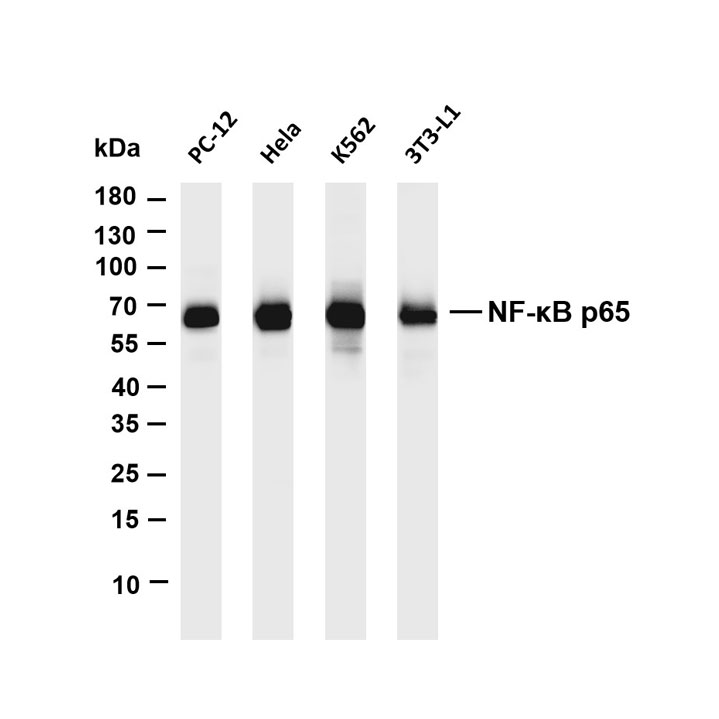
Core key proteins: DAF-2/DAF-16 (core nematode longevity proteins), SIR2 (yeast SIRT1 homolog), TOR (conserved aging regulator), FOXO (conserved transcription factor), SIRT1 (mammalian SIR2 homolog), AMPK (conserved energy-sensing kinase), insulin/IGF-1 receptor (cross-species), autophagy-related proteins (LC3, Beclin-1), HIF-1α (hypoxia adaptation protein), p53 (cross-species DNA damage responder).
Core key proteins: DAF-2/DAF-16 (core nematode longevity proteins), SIR2 (yeast SIRT1 homolog), TOR (conserved aging regulator), FOXO (conserved transcription factor), SIRT1 (mammalian SIR2 homolog), AMPK (conserved energy-sensing kinase), insulin/IGF-1 receptor (cross-species), autophagy-related proteins (LC3, Beclin-1), HIF-1α (hypoxia adaptation protein), p53 (cross-species DNA damage responder).
Product list
-
{{item.title}}{{item.react}}{{item.applicat}}
Product list
Product name
Reactivity
Application
Related Resource Links
Related Promotional Journal Downloads

Explore Our Recommended Popular Products
More products
30,000+ high- quality products available online
Primary Antibodies, Secondary Antibodies, mIHC Kits, ELISA Kits, Proteins, Molecular Biology Products,Cell Lines,Reagents ...
Contact Us